Abstract. Based on a principal component analysis of 47 published attempts to quantify hydrophobicity in terms of a single scale, we define a representation of the 20 amino acids as points in a 3-dimensional hydrophobicity space and display it by means of a minimal spanning tree. The dominant scale is found to be close to two scales derived from contact potentials.
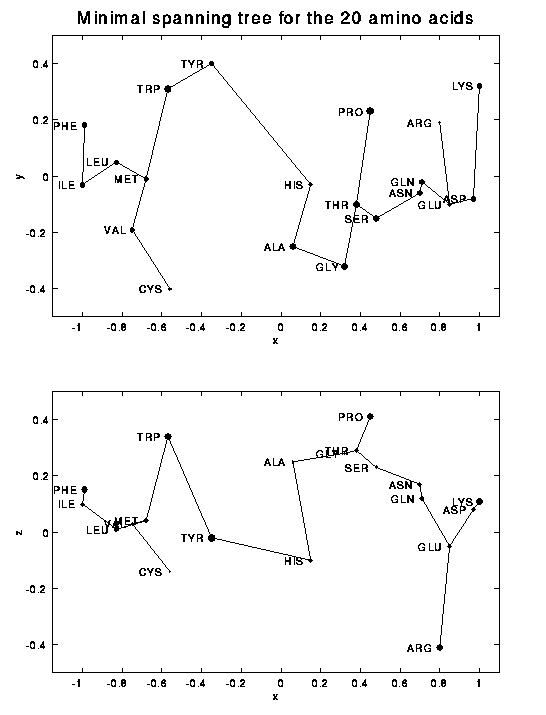
The topological structure of the minimal spanning tree is shown in the following figure. The tree is labelled twice, by the standard three letter and one letter abbreviations for the amino acids.

Acknowledgment. The authors gratefully acknowledge partial support of this research by the Austrian Fonds zur Förderung der wissenschaftlichen Forschung (FWF) under grant P11516-MAT.
The assignment of the amino acids to a quantitative hydrophobicity scale is a controversial problem. A review and evaluation of 46 different scales is given in the paper
J.L. Cornette, K.B. Cease, H. Margalit, J.L. Spouge, J.A. Berzofsky
and C. DeLisi,
Hydrophobicity scales and computational techniques for detecting
amphiphatic structures in proteins,
J. Mol. Biol. 195 (1987), 659-685.
Sometimes, different scales differ widely already in the order in
which the amino acids appear. This suggests that different scales
measure different properties more or less directly related to
hydrophobicity. The diversity of the scales then finds a natural
explanation in the fact that amino acids cannot naturally be ordered
in a linear way. However, a representation in higher-dimensional space
might be possible in such a way that close amino acids have similar
properties.
Restricting ourselves to those properties that are reflected in the
known hydrophobicity scales, we performed a
principal component analysis
(see also
here)
of 40 scales, namely the 39 complete scales from the above survey
(the 7 others are incomplete) and another scale (of so-called
`q-values') from
H. Li, C. Tang and N. Wingreen,
Nature of driving force for protein folding - a result from
analyzing the statistical potential,
Phys. Rev. Lett. 79, 765 (1997).
Keeping only the three dominant principal components, we found the
following coordinates of a three-dimensional representation of the 20
amino acids.
It is not surprising that the first (dominant) coordinate x represents
the bulk of the information in the 40 scales (75.7 percent). It can be
considered to be most closely related to the amount of polarity or
hydrophobicity, the common concept that all scales are supposed to
measure.
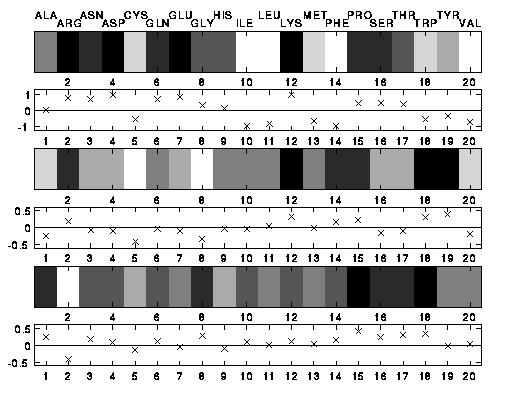
The following figure represents the three scales by grey level
bars (positive values are drawn dark, negative ones light) and in
additions by marking the levels with crosses.
The top scale contains x, the best compromise to a linear
hydrophobicity scale. (The numbers are the positions of the amino
acids in a lexicographic ordering.)
A
minimal spanning tree
analysis (see also
here)
reveals that the appropriate nearest
neighbor relation between amino acids is not fully linear. (The
missing third dimension is indicated by the size of the markers; the
fattest dots have a large positive missing coordinate.)
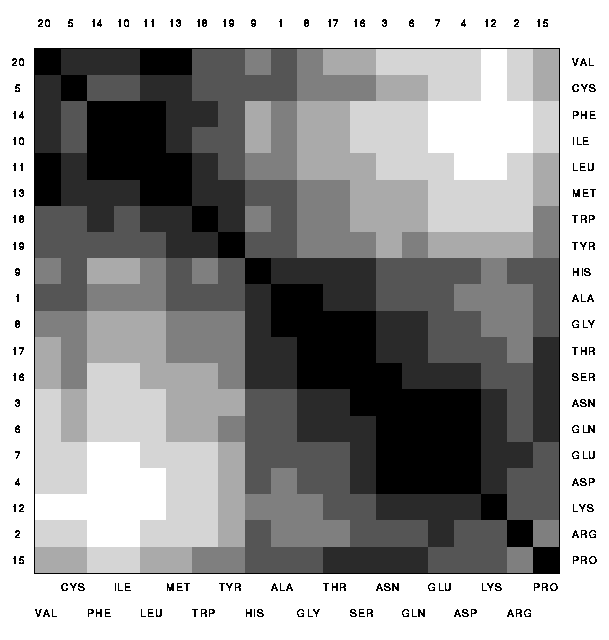
The deviation from a linear ordering can also be seen by looking at a
display of the distance matrix. Here the ordering has been chosen by
appending the branches of the topological tree at the closer ends of
the `backbone' of the tree. The distance is coded by grey values;
dark entries correspond to close pairs.
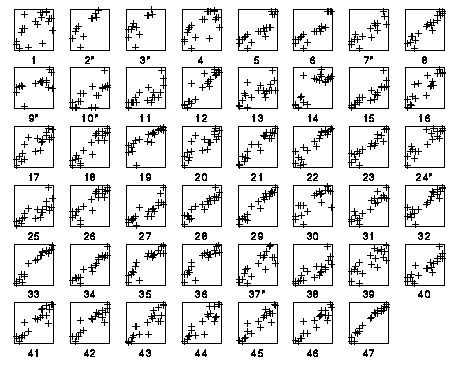
Finally we consider how close a linear transformation of the original
47 scales is able to approximate the dominant scale x found above.
We plotted each scale, linearly transformed to the range [-1,1],
against x. (The 7 incomplete scales not used in the principal
component analysis are marked with an asterisk$^*$.)
As one easily sees from the plot, the scale that gives the best
approximation to the dominant x scale (with a correlation of 0.982)
is scale 47, the scale by Tang et al.. The other
scales 1-46 correspond to Cornette et al.
according to the following list. (Among these, the scale closest to x
is scale 33=MIJER by Mijazawa and Jernigan, whose contact potentials
were also used in a different way for the derivation of the
Tang et al. scale.)
Note, however, that the correlation coefficient is a quite generous
measure of closeness of two scales. In particular, whether one
transforms the Tang et al. scale (whose correlation coefficient 0.982
with x is best) linearly such that either
(i) its mean and standard deviation agrees with x (see x' below) or
(ii) the extremal values are at -1 and 1 (see x'' below),
the scales don't match very closely:
ALA ARG ASN ASP CYS
.06 .80 .70 .97 -.56
-.25 .19 -.06 -.08 -.40
.25 -.41 .17 .08 -.14
GLN GLU GLY HIS ILE
.71 .85 .32 .15 -1.00
-.02 -.10 -.32 -.03 -.03
.12 -.05 .28 -.10 .10
LEU LYS MET PHE PRO
-.83 1.00 -.68 -.99 .45
.05 .32 -.01 .18 .23
.01 .11 .04 .15 .41
SER THR TRP TYR VAL
.48 .38 -.57 -.35 -.75
-.15 -.10 .31 .40 -.19
.23 .29 .34 -.02 .03
The precise procedure was as follows:
By a linear transformation we normalized each scale to mean 0 and
standard deviation 1, then computed the singular value decomposition
(also known as Karhunen-Loeve transform) and kept only the contributions
to the three dominant scales x, y, and z. (The degree of explanation
of the three scales, defined as the quotient of the corresponding squared singular value and the sum of squares of all singular values, was
75.7, 7.2, and 5.5 percent accounting for all but 11.6 percent of the
information in all 40 scales.)
By another linear transformation we shifted the scales such their range
was symmetric about zero, and multiplied all coordinates by the same
constant such that x ranges between -1 and 1. We then rounded the
values to two decimal places. (The large disagreement between the
various scales implies that there is no point in representing the
scales with higher accuracy; probably only the first figure is
significant.)
Thus we regard x as the best compromise to a linear hydrophobicity
scale. Polar amino acids have x>>0, hydrophobic ones have x<<0. Since
between -.35 (TYR) and .06 (ALA), there is a large gap in the possible
values for x, at least the classification into more or less polar
amino acids and more or less hydrophobic ones is unambiguous.
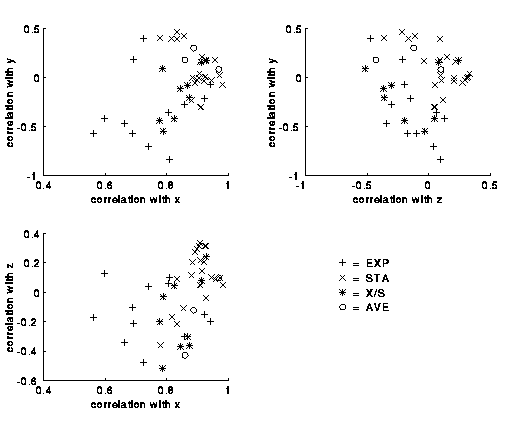
label Cornette code correlation with
x y z
1 EXP ZIMMR 0.60 -0.42 0.13
2 EXP N TAN 0.81 -0.84 0.10
3 EXP NTANR 0.74 -0.71 0.04
4 EXP JONES 0.69 -0.57 -0.10
5 X/S LEVIT 0.84 -0.12 -0.37
6 X/S HOPPW 0.87 -0.08 -0.30
7 EXP YUNGD 0.86 -0.28 -0.30
8 EXP FAUPL 0.94 -0.08 -0.20
9 EXP ZASLZ 0.56 -0.58 -0.18
10 EXP WOLF 0.72 0.40 -0.48
11 EXP KUNTZ 0.69 0.18 -0.22
12 EXP ABODR 0.92 -0.22 -0.15
13 EXP MEEK 0.66 -0.47 -0.35
14 EXP BULDG 0.81 -0.36 0.06
15 AVE EISEN 0.86 0.18 -0.43
16 AVE KYTDO 0.89 0.30 -0.12
17 STA CHOTH 0.86 0.42 -0.11
18 STA WERSC 0.92 -0.04 0.21
19 STA JANIN 0.83 0.39 0.09
20 STA OLSEN 0.82 0.40 -0.17
21 STA MEIRO 0.95 -0.03 0.10
22 X/S PONNU 0.93 0.17 0.24
23 STA NNEIG 0.92 0.21 0.15
24 STA ROBOS 0.88 -0.24 0.12
25 STA CHDLG 0.78 0.40 -0.36
26 STA WSDLG 0.88 -0.01 0.20
27 STA JADLG 0.83 0.46 -0.22
28 STA GUY 0.93 0.17 -0.04
29 AVE GUY M 0.970 0.08 0.10
30 X/S KRIDG 0.78 -0.44 -0.20
31 X/S KRIGK 0.91 0.15 0.08
32 STA NIOII 0.91 0.16 0.22
33 STA MIJER 0.973 0.02 0.10
34 STA ROSEF 0.96 0.18 0.09
35 STA SWEET 0.91 -0.30 0.05
36 STA SWEIG 0.91 -0.31 0.05
37 X/S REKKR 0.82 -0.42 0.05
38 X/S VHEBL 0.79 0.09 -0.52
39 X/S FROMM 0.79 -0.55 -0.03
40 X/S EIMCL 0.87 -0.21 -0.36
41 STA PRIFT 0.91 0.04 0.34
42 STA PRILS 0.91 -0.01 0.32
43 STA ALFT 0.89 -0.06 0.27
44 STA ALTLS 0.90 -0.03 0.29
45 STA TOTFT 0.93 0.01 0.31
46 STA TOTLS 0.92 0.00 0.32
47 TANG ET AL. 0.982 -0.08 0.05
Plotting the correlations for the various scales reveals a marked
difference between experimental (EXP) and statistical (STA) scales.
(Scales marked X/S are based on a mixture of experiment and statistics,
and scales marked AVE are averages of other scales.
The Tang et al. scale is marked STA.)
x x' x''
0.06 0.16 0.25
0.80 0.59 0.65
0.70 0.58 0.64
0.97 0.79 0.84
-0.56 -0.42 -0.30
0.71 0.72 0.78
0.85 0.84 0.89
0.32 0.48 0.55
0.15 0.23 0.32
-1.00 -0.93 -0.79
-0.83 -1.16 -1.00
1.00 0.96 1.00
-0.68 -0.68 -0.55
-0.99 -1.13 -0.98
0.45 0.45 0.52
0.48 0.63 0.69
0.38 0.44 0.51
-0.57 -0.55 -0.43
-0.35 -0.26 -0.15
-0.75 -0.62 -0.50
Molecular Modeling of Proteins
Arnold Neumaier (Arnold.Neumaier@univie.ac.at)